Highlight:
scNext is a generative foundation model that transforms static single-cell data into predictive temporal sequences, enabling the forecasting of cellular evolution, developmental potential, and long-term therapeutic responses.
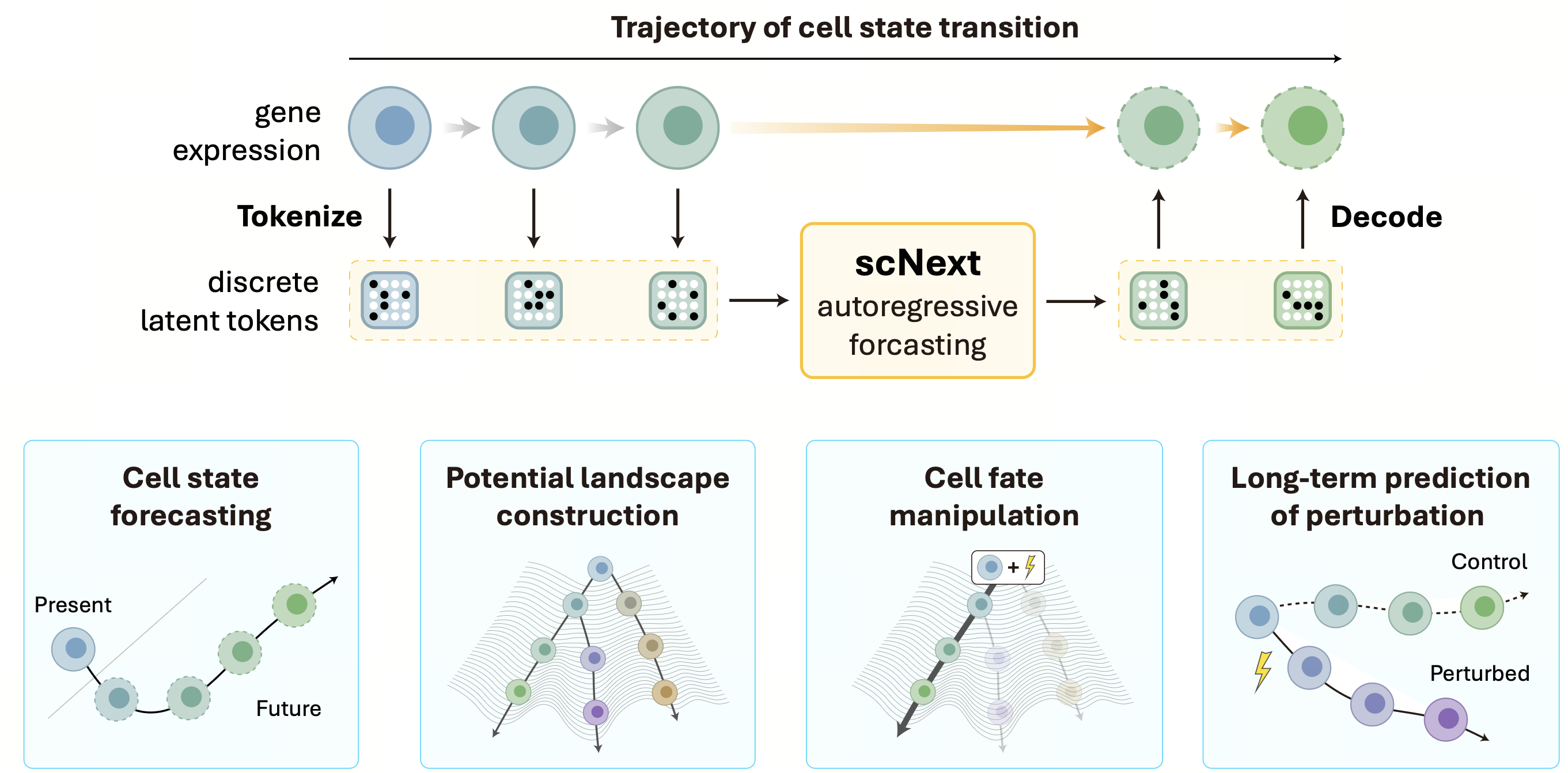
Single-cell sequencing has revolutionized our ability to profile cellular diversity, yet standard datasets capture only static molecular snapshots of inherently dynamic biological processes. To address this limitation, we introduce scNext, a temporal foundation model designed to learn and generate cellular trajectories directly from single-cell transcriptomic data.
Large-Scale Temporal Learning The development of scNext began with the construction of scBaseTraj, the first large-scale single-cell temporal dataset. By integrating RNA velocity, pseudotime estimates, and trajectory inference across 90 million cells, we converted static profiles into 48 million explicit multi-step cellular sequences. scNext utilizes a two-stage architecture: a vector-quantized variational autoencoder (VQ-VAE) discretizes high-dimensional gene expression into compact latent tokens, and an autoregressive transformer predicts the temporal evolution of these tokens. This discrete modeling approach significantly improves scalability and training efficiency compared to continuous gene-space models.
Forecasting Cellular Evolution and Potential Unlike traditional methods that retrospectively order cells along a pseudotime axis, scNextfunctions as a predictive engine. Starting from a single observed state, the model autoregressively generates plausible future trajectories, effectively simulating developmental progression. Furthermore, by quantifying the entropy of its predictive distribution, scNext infers a potential energy landscape that characterizes cellular plasticity. High-entropy states correspond to high developmental potential (e.g., stem cells), while low-entropy states indicate lineage commitment, allowing for the quantitative assessment of differentiation bias.
Predicting Therapeutic Perturbations A critical application of scNext is its ability to predict the long-term trajectory of cellular development following external perturbation. We demonstrated that the model can integrate drug molecule representations to forecast post-perturbation state transitions. For example, in hematopoietic stem cells treated with all-trans retinoic acid (ATRA), scNext correctly predicted a diversion from stemness toward myeloid and dendritic cell lineages. Similarly, in cancer cell lines treated with Lapatinib or Sodium Butyrate, the model accurately reproduced complex downstream effects, including pathway inhibition and cell-cycle arrest. By reframing single-cell modeling as a temporal forecasting problem, scNext offers a new computational paradigm for studying disease progression and simulating therapeutic interventions in silico.
