Research Philosophy
Our mission is to master the universal language of atomic interactions. By uniting expertise in physics, chemistry, biology, and computer science, we develop foundation models that decode the complexities of biomolecules at scale. We integrate powerful AI, advanced computing platforms, and high-throughput wet labs into a seamless feedback loop.
Here, real-world experimental data continuously refines our models—capturing the most vital insights for drug discovery. We pledge a steadfast investment in long-term fundamental research and will open-source all resulting frameworks to empower the community and accelerate the pace of discovery. Join us in this collective effort to push scientific frontiers and shape the future of drug discovery together.
Publications
-
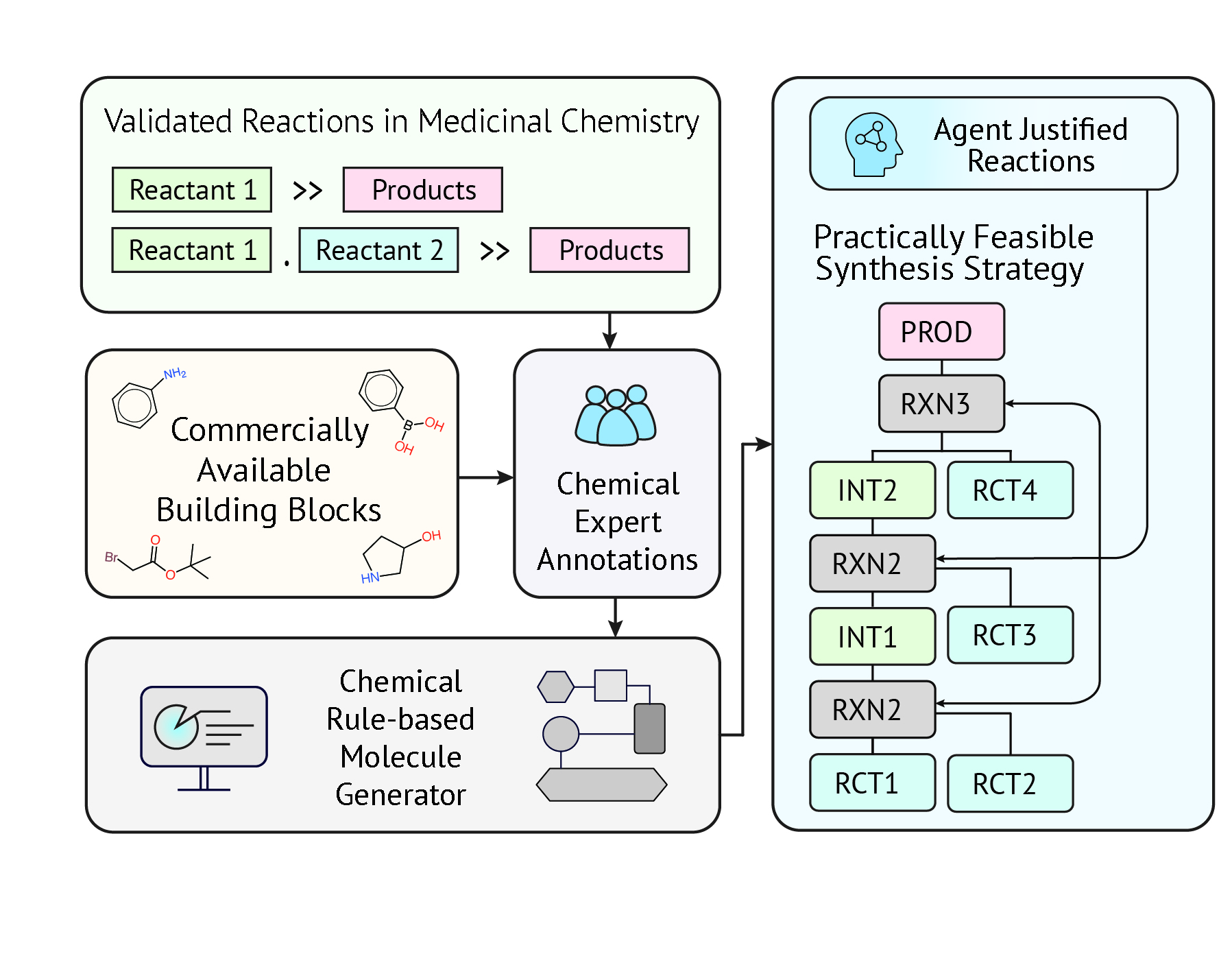
FeaSynth: Knowledge-Guided Synthesizable Projection for Feasible Synthesis Pathway Generation
Highlight: FeaSynth integrates an Expert Encoding Scheme with 145 annotated reaction templates to resolve critical chemical conflicts and significantly outperform SOTA baselines in practical synthesizability. We also establishes a standardized Reaction Pathway Feasibility (RPF) Score to rigorously quantify route viability and ensure only the most robust, experimentally valid pathways are selected.
-
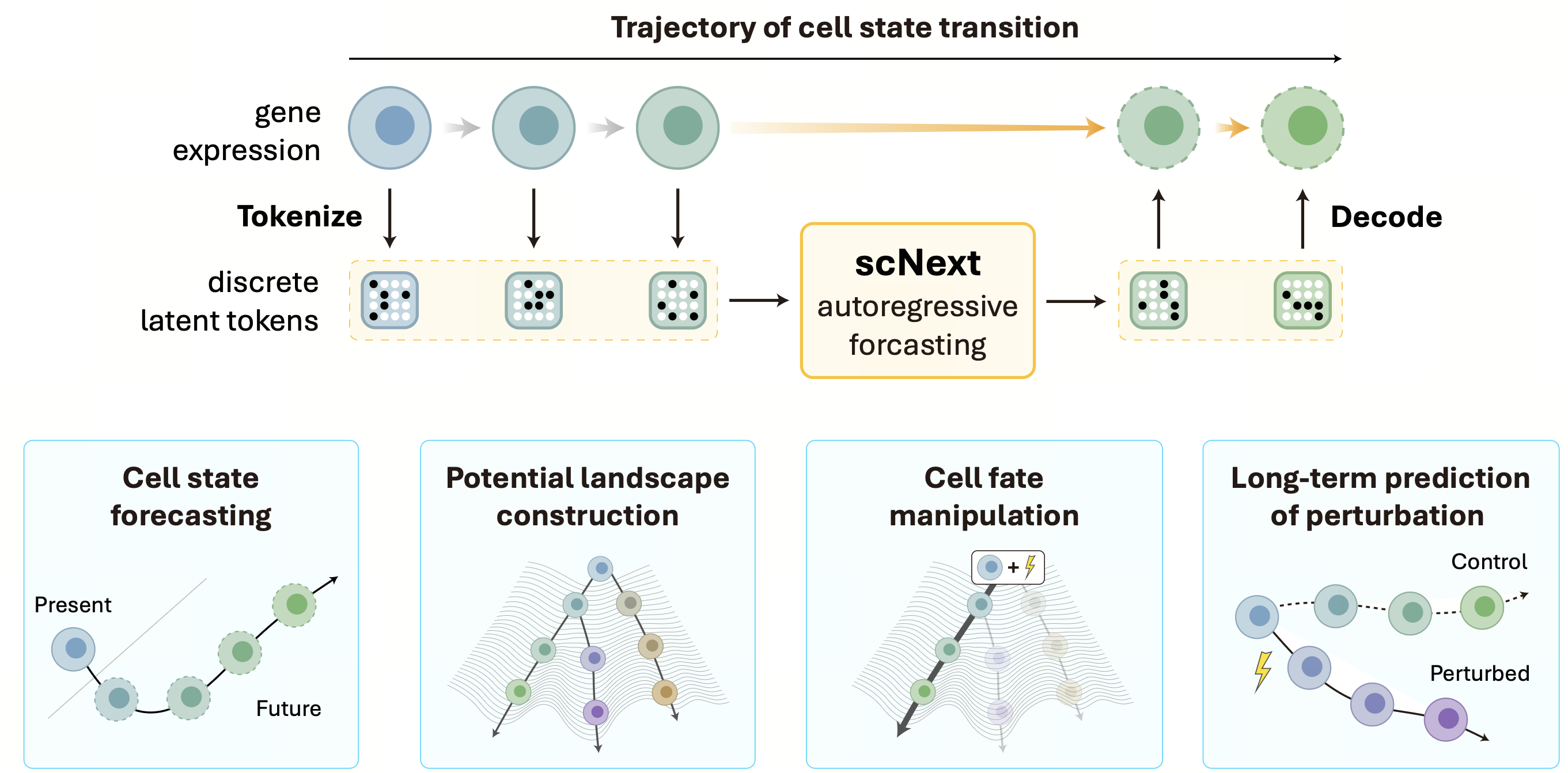
scNext: A Foundation Model for Forecasting Cellular Dynamics and Perturbation Responses
Highlight: scNext is a generative foundation model that transforms static single-cell data into predictive temporal sequences, enabling the forecasting of cellular evolution, developmental potential, and long-term therapeutic responses.
-
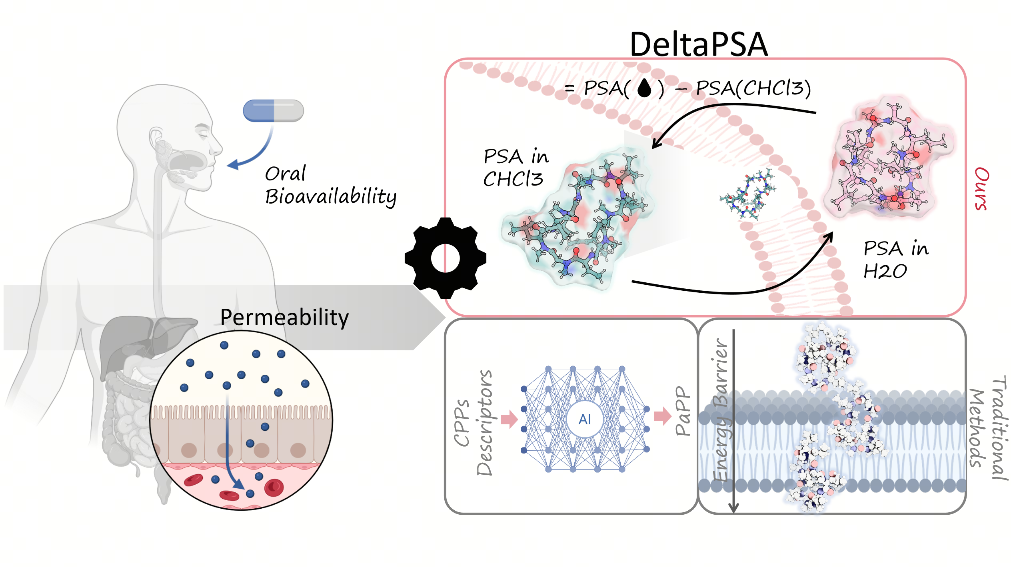
Delta PSA: A New Metric for Conformational Dynamics Underlying Macrocyclic Peptide Permeability
Highlight: Cost-Effective Prediction of Macrocyclic Peptide Permeability via Dynamic Conformational Analysis We introduce a computationally efficient molecular dynamics framework that accurately predicts macrocyclic peptide permeability by quantifying environment-dependent conformational adaptability and specific shifts in polar surface area .
-
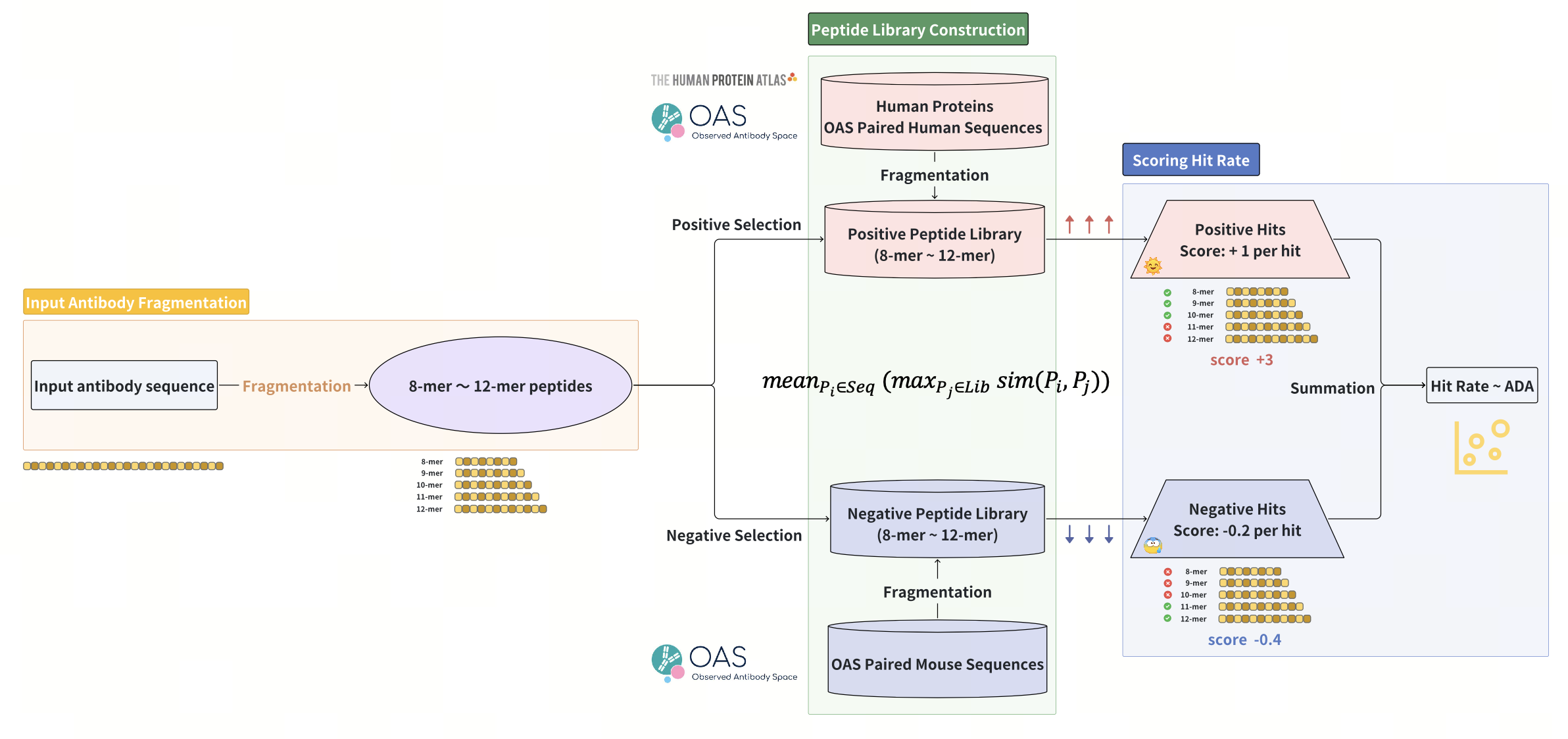
Antibody immunogenicity prediction and optimization with ImmunoSeq
Highlight: This study presents ImmunoSeq, an interpretable and applicable method for immunogenicity prediction. ImmunoSeq demonstrated superior ADA correlation and humanness classification accuracy compared to deep learning models, while accurately predicts ADA reductions in humanization, enabling sufficient sequence optimization for humanness.
-
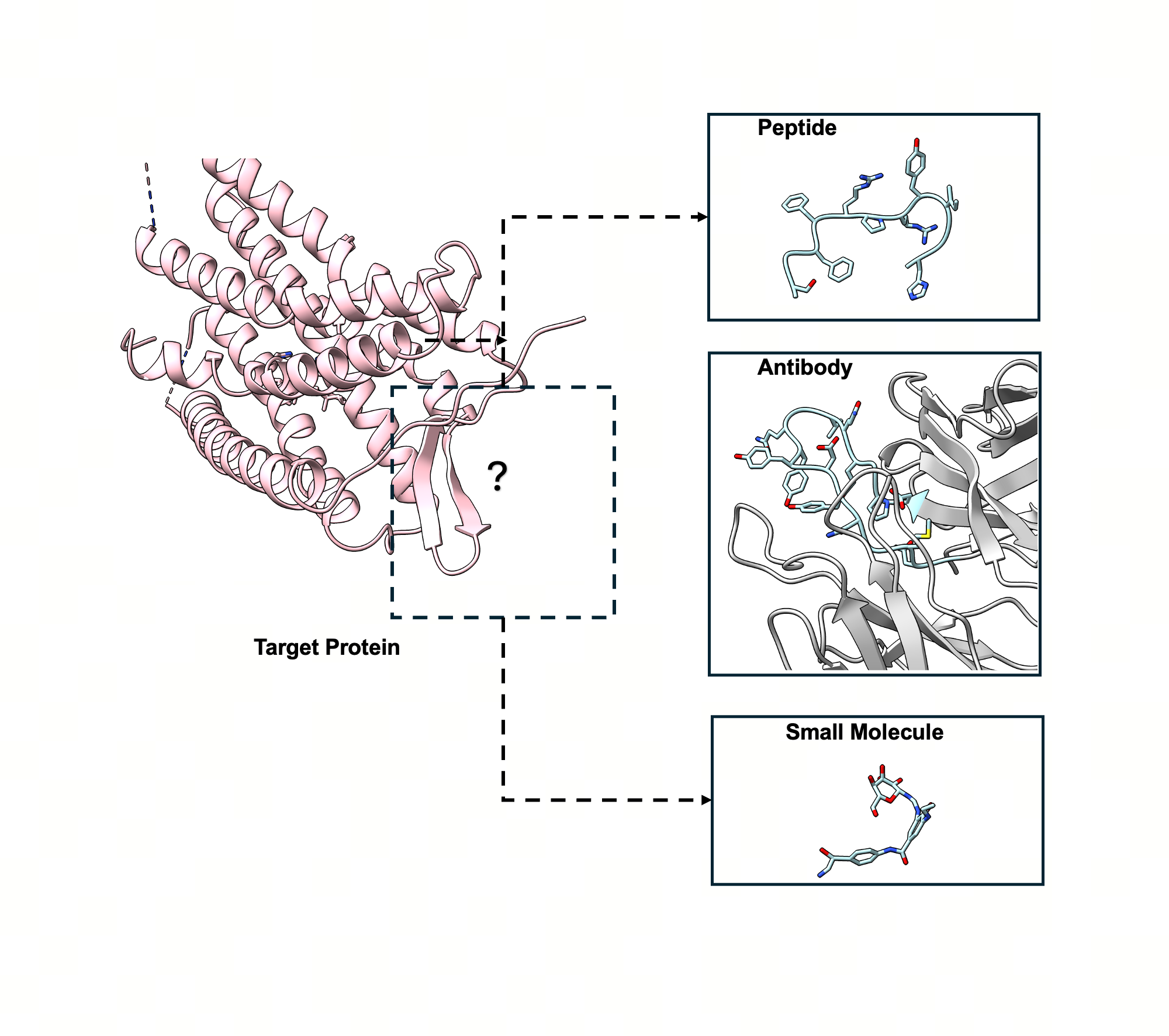
UniMoMo: Unified Generative Modeling of 3D Molecules for De Novo Binder Design
Highlight: UniMoMo is the first all-atom geometric latent diffusion framework capable of designing peptides, antibodies, and small molecules within a unified generative model, demonstrating superior performance and cross-domain knowledge transfer in de novo binder design.
-
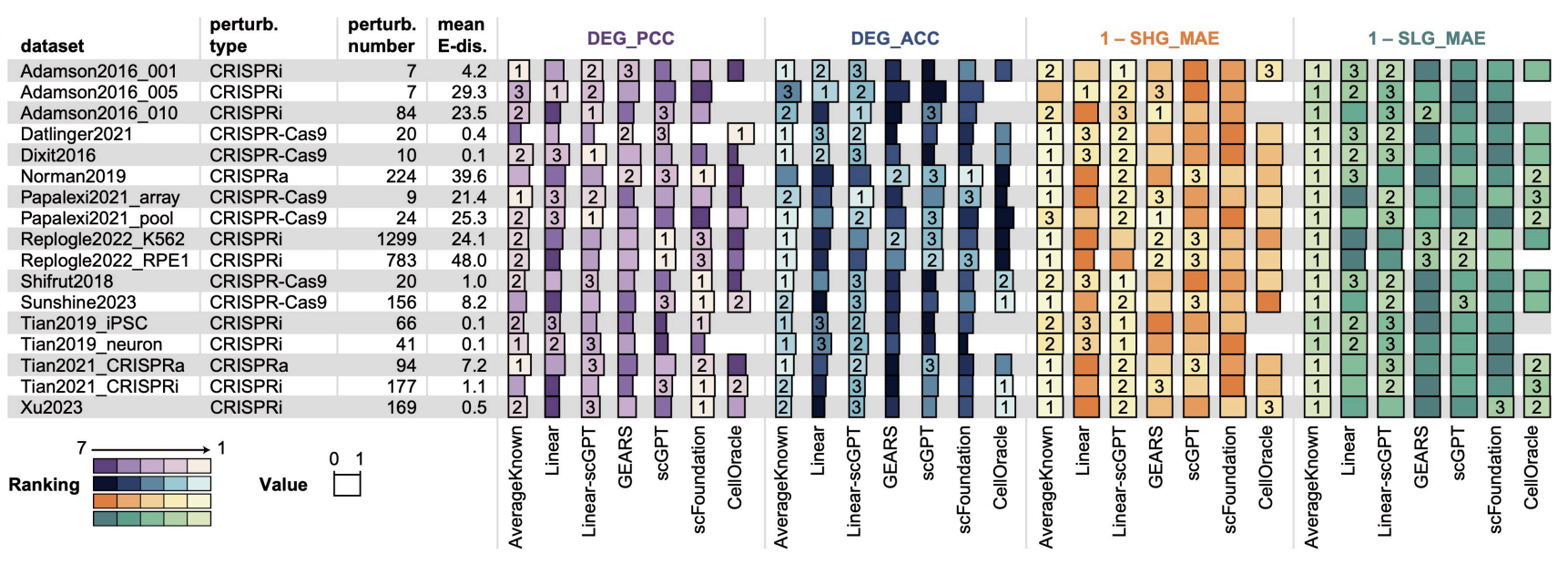
Benchmarking AI Models for In Silico Gene Perturbation of Cells
Highlight: This study establishes a comprehensive benchmarking framework for in silico gene perturbation, systematically evaluating ten AI methods across four biological scenarios to standardize assessment and advance computational drug discovery.